Introduction
It has been reported that in cells the predominant DNA conformation is in its B form (Right handed), which is known to play a pivotal role in cellular processes. These include carrying of genetic information thereby playing a role in Protein synthesis.1 DNA molecule is constantly insulted by several chemicals, metals, and various environmental factors and studies including our group have revealed that metals are capable of interacting with proteins and DNA causing changes in the structure and oxidative deterioration. Different conformations of DNA are known to exist as B, Z, C and ψ DNA. It is also known from literature that in these different conformations, the double helical geometry, major and minor groove depth and width are varied. 2 The promoter regions are GC rich and over-represented in the eukaryotic genome that forms hairpin secondary structures, and dinucleotide repeats which are shown to be unstable and polymorphic. 3, 4 The Poly (GC) sequence are unique class of repeats with alternating purines and pyrimidines form secondary structures, particularly hairpin loops that alter DNA replication and repair. 5 Under specific conditions, the Poly (GC)) adopts a Z DNA conformation (left handed) indicating its several roles in cellular processes. 6, 7, 8, 9, 10 and lead to different neurological disorders lead Alzheimer’s disease (AD) and Huntington disease (HD). Extensive chemical and biochemical studies evidenced binding of metals to DNA which is known to influence numerous biological processes like replication and transcription. A high level of gene expression is seen in brain cells, which can be altered due to DNA damage and affects the transcription processes and subsequently conformational variations in DNA. 11 Studies also reveal that (Aβ) and tau undergo conformational changes on interaction with Cu invitro.12, 13 Binding of metals and (Aβ) to DNA results in conformational changes from the B DNA form to an altered B DNA and ψ –DNA, which is the transition that occurs in the pathology of AD. 14 This change alters the normal functions of the cell which enhances the DNA instability, and is thus implicated in the cellular dysfunctions in brain disorders. 15, 16, 17, 18 The Aβ peptide interacts with Cu and can initiate peptide misfolds and aggregation. 19 As limited has been reported with respect to conformational changes of DNA on binding to Cu and Aβ, which will provide clues as to how they are involved in neurodegenerative disorders.
Materials and Methods
Poly (GC) Sample
The Synthetic double-stranded DNA polymer, Poly GC sodium salt with Lot No 31K1442 was obtained from Sigma Aldrich. It was dissolved in Milli Q water without further purification. DNA dilutions were prepared from the stock, by diluting to the desired concentrations using Tris-HCl (5mM strength, 7.4 pH) buffer for UV and CD studies.
Aβ 1-16 peptide
The 16 amino acid residue of Aβ with sequence DAEFRHDSGYEVHHQK with 1956 Da as molecular weight was procured from M/s USV Peptides
Copper chloride (CuCl2.2H2O)
The CuCl2.2H2O was procured from Merck Schuchard. 50mM stock solution was prepared by dissolving 9.5 mg of CuCl2.2H2O in 1.1 ml of Milli-Q water. Further dilutions were used as and when required for experimentation.
UV/VIS absorption studies
Electronic absorption and thermal denaturation studies of Poly GC with and without copper chloride were done using Jasco V-530 spectrophotometer (Jasco, Japan). This was equipped with a Peltier temperature controller. Samples of Poly (GC) (1.3ug/ul) were dissolved in Tris-HCl buffer to arrive at a concentration of 5mM and pH of 7.4. Spectra of native DNA and complexes with Copper and Aβ were recorded in 1 cm path length cuvettes at wavelength range of 220nm and 320nm.
UV-thermal melting studies (absorbance Vs temperature) of Poly (GC) incubated in HEPES buffer (10mM and pH 7.4) with and without copper(1µM to 1mM). were recorded at 260 nm at temperature range between 25 to 90oC, (heating rate of 1.0°C/min). For each recording, the melting temperature (Tm) of Poly (GC) was determined as the transition midpoint.
Circular Dichroism studies
Changes in poly GC conformation were recorded in a Jasco J-715 spectropolarimeter at 25 °C with a 1 mm path length quartz cuvette. Readings were taken at 1nm intervels between 200nm and 320nm wavelength. The spectra were recorded with an average of four repetitive scans to improve the signal-to-noise ratio in each concentration.
A 1.3 µg/µl concentration of Poly (GC) in 5mM Tris–HCl buffer of pH 7.4 at concentrations ranging from 100µM to 1mM was studied with and without copper between 200 nm and 320 nm wavelength range and temperature range of 20 to 100°C at heating rate interval of 10°C.
Fluorescence studies
Fluorescence spectroscopy is determines structure and dynamics of nucleic acids. Fluorescence emission studies were done using a 1:1 concentration of Poly (GC) repeats and EtBr. The EtBr binding pattern of Poly (GC) was studied and the effect of Cu in different concentrations between 500 nm to 500 µM on the EtBr fluorescence was analyzed. Exitation of DNA/EtBr solutions were done at 530 nm, and spectra read from 550 nm to 650 nm using Jasco J-600 spectrofluorimeter.
Docking studies
Molecular docking was carried out to predict the determinants of binding through binding affinities via scoring functions. The primary objective was to define interactions of copper with nucleotide bases. A duplex of a model for Poly (GC) sequence was retrieved (PDB ID: 2ANA) from the Protein data bank. The docking studies were carried out with the crystal structure of Z-DNA dodecamer d (CGCGCGCGCGCG)2 of 0.75 Å resolution (PDB ID 4OCB) with Abeta 1-16 peptide for DNA- peptide interaction studies. Minimization of retrieved structures was done using conjugate gradient protocol along with CHARMM force field. This was done to depreciate ligands with Max Steps-2000 incorporated in the Discovery Studio 3.5 software.
Results
Interaction of Poly (GC) sequence with Cu and Aβ1-16 peptide
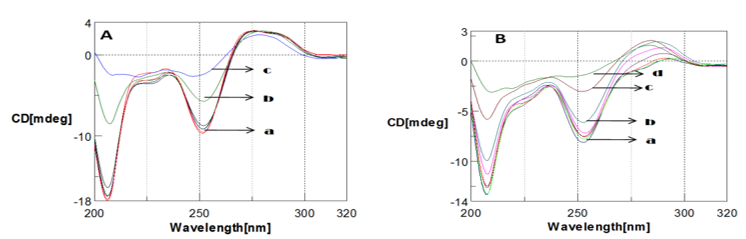
The effect of copper (Cu) and Aβ1-16 peptide on the Poly (GC) is shown in Figure 1, Figure 2, Figure 3. CD spectra of Poly (GC) indicate B-DNA conformation with a positive peak observed at 277 nm and a negative peak observed at 252 nm. This may be attributed to base stacking and right-handed helix respectively.20, 21 Changes in the positive band with an evident redshift to 298 nm (21nm) and narrow changes in the negative band with the visible redshift to at 255 nm and 209 nm as shown in Figure 1 were noticed upon addition of Cu from 100 µM to 250 µM. The decrease in the positive band with narrow changes in the negative band at 255 nm exhibited a C-like conformation of DNA. 22, 23 When the Cu concentration is further increased from 500 µM to 1mM, the positive band increased with a shift to 284 nm and the negative band decreased with a redshift to 258 nm and 209 nm. A C-DNA to altered B-DNA conformational change was observed which is depicted in Figure 2. It is thus revealed from the experiment that the DNA sequences have undergone a shift from the B DNA to C DNA conformation and C DNA to altered B DNA on binding to Cu.
To study the interaction, CD spectra of Poly (GC) sequence with and without Aβ1-16 peptide was recorded. B-form of DNA with typical bonds at 277 nm and 252 nm is shown in Figure 3. The magnitude of both the positive and negative bands diminished with a narrow shift in both the bands is with Aβ1-16 peptide due to DNA damage and conformational change. We also measured the Aβ1-16-DNA complex by varying the time, 2 hours, 24 hours, 48 hours and 72 hours is shown in Figure 4. Remarkable spectral variations at 208 nm were noticed due to the secondary structural changes in DNA. Further, studies with varied pH, the poly GC sequence remains sensitive to various pH values, structural changes were noticed upon varying the pH ranging from pH.5 to pH 7.4 are depicted in Figure 5. CD spectra indicate the higher DNA damage at lower pH values at pH 5 and pH 6.24 Poly (GC) sequences are sensitive to the different concentration of NaCl is shown in Figure 6. A decrease in the negative band at 252nm with complete disappearance of the negative band at 208nm indicating that salt-induced conformational changes are highly cooperative and Cu do not have much effect on the DNA-NaCl complex.
Individually, the Aβ1-16 peptide and Cu is known to induce conformational changes in DNA. The spectral variations observed may be because of the binding of Cu and Aβ1-16 peptide to a particular site, and also to accommodate the incoming Cu and Aβ1-16 peptide. This may allow the strands the DNA to be pushed apart resulting in widening of the major groove. This spectral shift along with the magnitude decrease of the positive and negative bands may be indicative of conformational shift during incubation with Aβ, resulting in disturbances in structure of DNA. This damage in turn may be attributed to change in winding angle of the helix due to super helical tension by Aβ1-16 and Cu.25 The interaction of Cu with DNA causes disturbances to the B-DNA. Change in the longer wavelength positive band of DNA is a change in DNA secondary conformation from B-conformation to C-conformation is involved with an increase in the winding angle. Together with the change in winding angle there are concomitant changes in the tilt of the base pairs and the distance of the base pairs from the helix axis, geometric changes. An increased degree of tilt of the basepairs, the chain affecting sugar puckering from anti to syn stacking pattern in the DNA structures results in the unwinding of the helices, that could be responsible for the C-DNA conformation. 26 The increase in the positive band and decrease in the negative band with the visible redshift to 256 nm is evidenced with higher concentrations of Cu. The increase in the positive band caused by superhelical tension due to the binding of Cu those results in a slight decrease in the winding angle of the base pairs leads to altered B-DNA conformation from C-DNA. The results indicate the interaction of copper –peptide complex on the base stacking thereby initiating DNA damage. Taken together, these changes indicate a more progressive DNA damage during the incubation with Aβ1-16 peptide and the secondary conformation of DNA was perturbed is shown in Figure 4. The binding of Aβ1-16 peptide and Cu with DNA pushes apart the helices and cause disturbance to B-DNA due to altered base stacking. The altered stacking of bases in each DNA strand results in unwinding of the helix.The extent of the twisting gives rise to variations among the DNA secondary structures that cause a decrease in the right handedness that altered B-DNA conformation.
Earlier reports indicated that Aβ, tau, and metals are capable of interacting with nuclear proteins and DNA causing DNA damage and conformational change that may cause changes in gene expression.27, 28 The current report provides a hint on the interaction of Aβ1-16 and Cu with Poly (GC) sequence DNA thereby altering DNA conformation.
Binding of Cu and Aβ1-16 peptide on the conformation of Poly (GC) sequence via UV Absorption
The most commonly used method to analyse the interaction of metals and proteins with DNA is UV-Vis absorption studies. Interactions of Cu and Aβ peptide with Poly (GC) sequence were done through UV absorption studies and the spectrum is shown in Figure 7. The spectra showed the absorption maximum at 254 nm, with a small change in absorbance when low concentrations of Cu was added to the DNA. An increase in absorbance with 100μM of Cu and an increase in absorbance with a peak shift to 250 nm (Blueshift) indicated structural changes of DNA due to the interaction of Cu. The peak at 254 nm could be attributed to the spin-allowed π–π* transitions of DNA. This change in shift also reveals the interaction of Cu with DNA via electrostatic interactions resulting in perturbations and disruption of secondary structure thus resulting in unwinding of DNA double helix to accommodate the incoming Cu. The Absorption spectra of Poly (GC) sequences were performed with varying concentrations of Amyloid β 1-16 is shown in Figure 8 . The spectra shows hypochromism upon increasing the concentration of Aβ, indicating that the Aβ interacts with DNA via an intercalative mode and strong stacking interaction between Aβ peptide and base pairs of DNA thus results in electronic perturbations that causes a twist in the double helix in DNA to recieve the incoming Aβ peptide.29, 30
Effect of Cu on the conformation of Poly (GC) sequences via Fluorescence studies
Similar to CD and UV absorption, Fluorescence studies were done to look at the interaction of Cu with DNA. The fluorescence spectra of Poly (GC) - EtBr complex in the absence and presence of Cu was recorded and is depicted in Figure 9. Poly (GC) complex shows the fluorescence maximum at 590 nm and the fluorescence intensity decreased remarkably with the increase in the concentration of Cu. The reduction in the fluorescence may be due to the quenching action of Cu with DNA-EtBr complex. The percentage of quenching was raised from 2.89% to 55% with an increase in Cu concentration. Cu preferentially bind to purine bases, mostly guanine, by forming covalent coordination bonds to the nucleophilic N7 atoms and change the conformation of DNA. 31, 32 This data indicates the competition of Cu with EtBr for occupying similar binding sites on Poly (GC) nucleotide sequence by displacing EtBr. Hence, the molecular conformation of the nucleotide sequence was affected due to Cu binding and the process of quenching will address the destabilization of DNA.
Effect of temperature on changes in conformation of Poly (GC) sequence
The temperature affects altering the stability of the double-stranded DNA and altered by the presence of metals and small molecules. To study the effect of temperature on Poly (GC) sequence DNA stability, CD spectra of Poly (GC) sequence with and without Cu from 20ºC to 90ºC was recorded. The spectra indicate characteristic of B-DNA is shown in Figure 10. The poly GC sequence is highly stable up to 80°C, showing a narrow change in both the bands and a decrease in CD bands starts from 80ºC and 90ºC, with a loss of intensity of the negative band at 252 nm and narrow changes in the positive band with a redshift to 280 nm. The decrease in the negative band and redshift is due to the changes of the base-paired structure with increased temperature. Further, in the presence of 100 µM Cu changes in conformation with the positive band shift to 291nm were observed and presented in Figure 10. Minor changes in the negative band and decrease in the positive band were observed up to a temperature of 60°C. These changes resembled the C-form of DNA as the DNA-Cu complex. Further, as temperature increased, there was an increase in the intensity of the positive band with a redshift to 290 nm was noticed to at 90ºC. The results revealed that both the bonds are sensitive to temperature and there is a difference between the kinetics of native Poly (GC) and with Cu (100 µM). The Poly (GC) sequence is stable up to 80ºC, and with 100 µM Cu it changes to 70ºC. The UV melting profiles shown in Figure 10 provides details of the physical attributes of poly GC sequence. At high temperature (about 80ºC) cross-strand hydrogen bonds found between complementary DNA strands may be broken, and disrupts the conformation giving rise to a pre-melted stable conformation. Transition temperature varies with binding of Cu to the bases of DNA, which inturn lowers base pairing and stacking and breaking of hydrogen bonds, loosening of base-base interactions and tilting of the bases helps unwinding the helix thereby destabilizing it. Cu has a very higher affinity towards G-C base pairs and thus they destabilize the double helix and decrease the Tm values to 70ºC by inducing unfolding due to DNA damage leads to a structural change of the DNA. Hence, the Cu binding to DNA affects the thermal stability of the Poly (GC) sequence. These results are comparable with the melting profiles of UV studies, which show the Tm of Poly(GC) is 83ºC, with 100 and 250 µM Cu it is 77ºC. The melting of strands™ depends on the percentage of GC base pairs. Tm is known to increase when there is an increase in the GC bases 33, 34 corroborates with our results. Cu has a greater affinity for heteroatoms on the bases and binding of this with bases of Poly (GC) sequence results in breakage of hydrogen bonds that hold G-C bases of the complementary strands of DNA. The DNA-copper complex is very sensitive to temperature variations and is complex with respect to the binding sites between the strands which will then compete for the binding site resulting in disruption of the secondary structure. This inturn results in a decrease in the Tm values. 35 The Tm data suggest that the poly GC sequence is sensitive to higher temperatures and copper concentrations wherein Cu binds to phosphate and nucleotide of DNA and unwinds the strands at a higher temperature and eventually lowers its stability.
Molecular docking studies
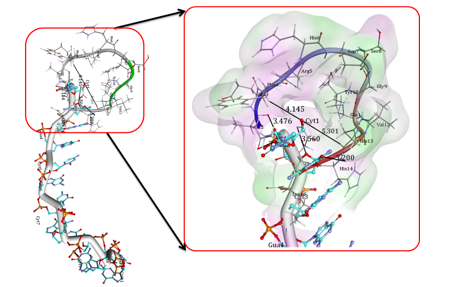
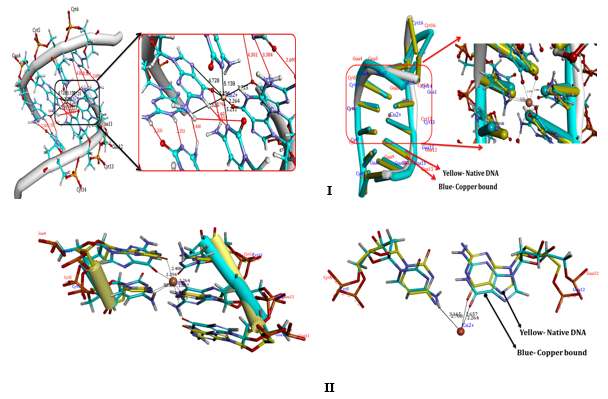
Docking studies provide some insight into the interaction between copper and Poly (GC) sequence, which can substantiate the experimental results. Figure 11 I, II show the docking conformation of DNA with Cu. Our docking studies indicate that Cu on binding to DNA, forms covalent mode of DNA binding across Guanine and cytosine, particularly N7, 06 of Guanine of A chain and N3 of Cytosine residue of B chain. The coordination of N7 is expected as N7 atoms of purines are the most electron-dense and will be accessible sites in DNA. These are displayed in the major groove of the double helix. The coordination of Cu with four nucleotide residues indicates the strong distortion of the double helix leading to altered conformations. The docking interaction of the Aβ1-16 peptide with the crystal structure of the d(CGCGCGCGCGCG)2 dodecameric duplex ((PDB ID 4OCB) is given in Figure 12 and exhibits parallel duplexes arranged aslong helices packed in a hexagonal fashion, similar to all other Z-DNA oligomer structures. Docking results indicate strong interaction of critical residues of Aβ(1-16) like ALA2, GLU3, HIS13, HIS14, and GLN15 with Cytosine and Guanine nucleotide of DNA as indicated in Figure 12. Hence, from In silico docking results, we conclude that there is a strong binding affinity of A beta peptide across the Poly GpG repeats of DNA, which induces the conformational change and declines the stability of the DNA.
Further, our insilico results confirm the disturbance of B-DNA. This may lead to the alteration of the base stacking in each of the strands which results in the change of conformation of DNA to an altered B-DNA form. Thus, our Insilco results corroborate with the invitro studies of DNA binding which could change the secondary and tertiary structure of DNA resulting in its destabilization.
Figure 1
CD spectra of Poly (GC) with varied concentration of copper in 5mM Tris-HCl buffer., a. Poly (GC) alone, b. Poly (GC) +100 µM CuCl2, c. Poly (GC) +250 µM CuCl2.
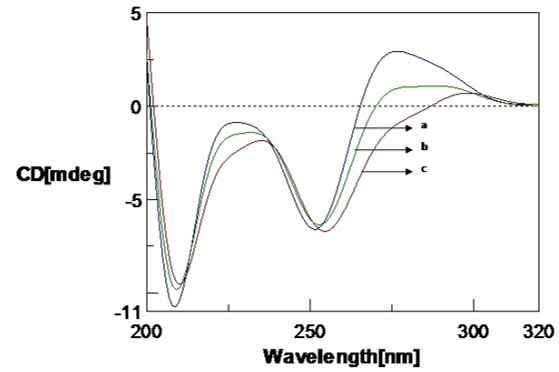
Figure 2
CD spectra of Poly (GC) with varied concentration of copper in 5mM Tris-HCl buffer. a. Poly (GC), b. Poly (GC) +500 µM CuCl2, c. 1mM CuCl2.
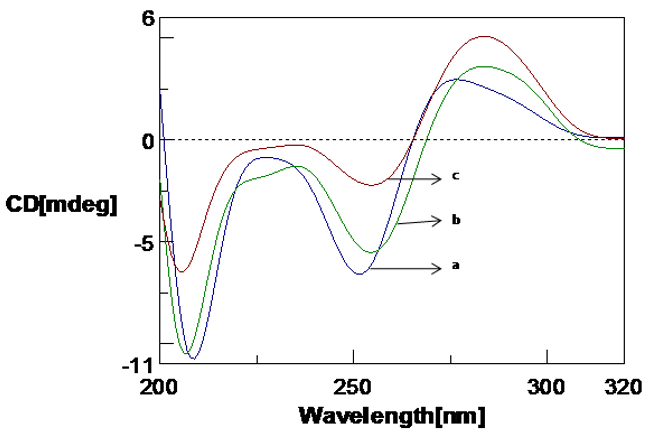
Figure 3
CD spectra of Poly (GC) with Amyloid β(1-16) peptide a. Amyloid β(1-16) peptide b. Poly (GC) sequence, c. Poly (GC) sequence with Amyloid β(1-16) peptide( 10µM)
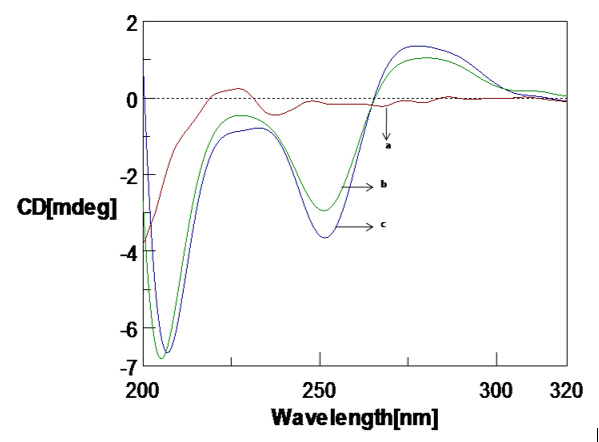
Figure 4
CD spectra of Poly (GC) sequence with Aβ1-16 and with varying time. a. Poly (GC), b. Poly (GC) +Aβ1-16, c. Poly (GC) +Aβ1-16 (24 hrs), d. Poly (GC) +Aβ1-16 (48 hrs), e. Poly (GC) +Aβ1-16 (72 hrs)
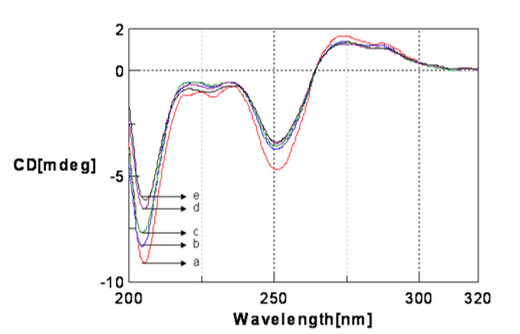
Figure 5
CD spectra Poly (GC)) sequence with varied pH in 5mM Tris-HCl buffer. a. Poly (GC) sequence pH.7.4, a. Poly (GC) sequence pH 6, c. Poly (GC) sequence pH 5.
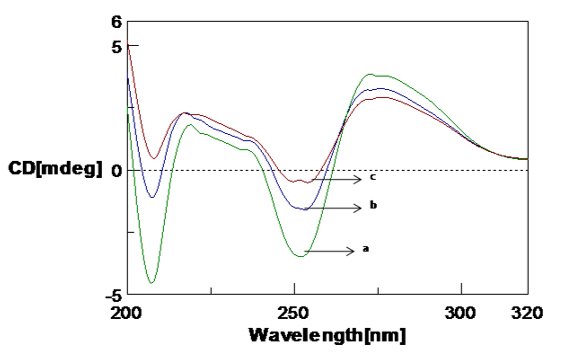
Figure 6
CD spectra showing the effect of NaCl on Poly (GC) in 5mM Tris-HCl buffer. a. Poly GC, b. Poly (GC) +200 µM NaCl.
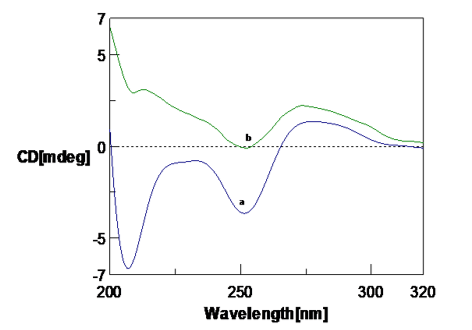
Figure 7
UV absorption spectra of Poly (GC) sequence with copper chloride in 5mM Tris-HCl buffer (pH.7.4) a. Poly (GC), b. Poly (GC) +10μM CuCl2, c. Poly (GC) +50μM CuCl2.
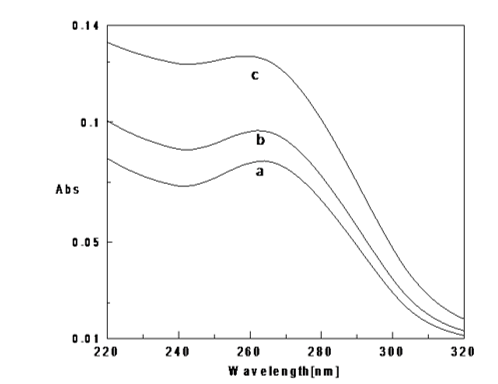
Figure 8
UV absorption spectra of Poly (GC) sequence and with Aβ1-16 peptide in 5mM Tris-HCl buffer (pH.7.4) a. Poly (GC), b. Poly (GC) +Aβ1-16(10μM) c. Poly (GC) +Aβ1-16(20μM)
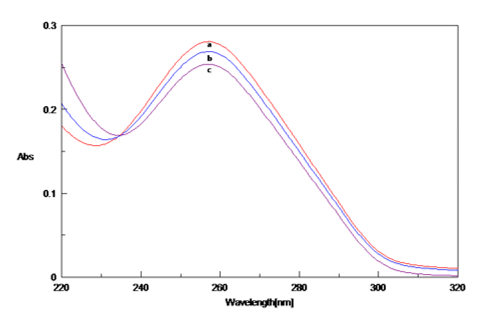
Figure 9
Fluorescence emission spectra of Poly (GC) -EtBr complex in the presence of varied concentrations of CuCl2.a. Poly (GC),b. Poly (GC) +100µM CuCl2 ,c. Poly (GC) +250µM CuCl2,d. Poly (GC) +500µM CuCl2
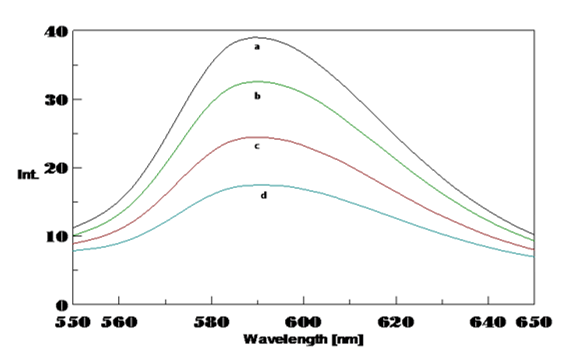
Figure 10
CD spectra of the effect of temperature on Poly (GC) sequence. A, a. Poly (GC) with varying temperature from 20- 70°C ( Minor changes), b. Poly (GC) with a temperature of 80°C, c. Poly (GC) with a temperature of 90°C
Poly (GC) with 100 µM CuCl2 with varying temperature, a. From 30- 60°C, b. 80°C, c. 90°C( spectra of 100°C is not shown).
Figure 11
Docking interactions of Copper, with Poly (GC). Docking interactions of Copper, with poly Poly (GC) (PDB ID:2ANA) showing coordination of copper across N7,06 of Guanine and N3 of Cytosine nucleotide bases. B) The nucleotide bases showing the orientation of nucleotide base pairs/significant rotations across the base pairs and Unbound Copper forms of DNA helices (Blue) indicating the shift in the position upon binding of the Cu representing the changes in the altered B-DNA conformations.
Discussion
A high level of gene expression is observed in brain cells. Cells in regions of the brain that are very sensitive to changes to DNA damage include the basal ganglia and cortex. The predominant conformation of the DNA is in its B DNA form. Biological functions of DNA are correlated with the molecular structure. Changes in the conformation may lead to alterations in a number of biological functions.36, 37
Cu is important for many metabolic processes in cellular functions including CNS development. However, Cu also plays a vital role in AD by influencing oligomerization and aggregation of Aβ peptides. Compared to other tissues in the body, the brain is more sensitive to oxidative damage and the effects of the DNA change is specifically seen in the basal ganglia and parts of the cortex. The accumulation of Cu exhibits toxicity due to the interaction with DNA and proteins.38, 39 The studies revealed that increased concentration of Cu in the brain alters DNA stability and biochemical functions of DNA. Some hypothesis has been proposed to explain the toxicity and the role of Aβ peptides that may alter the gene expression at promoter regions in the genome and lead to brain pathology and in fact, Cu has been considered as an important etiological factor for neuro degenerative diseases.40
GC-rich sequences are the major components of the promoter region and are involved in gene expression. These are commonly seen in the 5' regions of AD specific genes like PS1, PS2, and APOE.41 The DNA structural property and duplex stability is an important factor for normal DNA functions. This study is evidenced that Cu interacts with DNA and causes a conformational change that results in a decrease in the positive long-wavelength band involved in secondary and tertiary structural changes. The origin of the structural changes in the long-wavelength positive band of DNA is indicated as a change in DNA secondary conformation from B-conformation to C-DNA conformation. Winding angle increase of the base pairs may lead to conformational change. C-DNA conformation may be regarded as a distorted B-conformation. 42 Cu has strong affinity to bases and bind to specifically to G-C (N7 of guanine and N3 of cytosine) in poly(dG-dC). poly(dG-dC) sequences which are confirmed by docking studies depicted in fig.8A, B. As a consequence of the interaction of Cu with N7 nitrogen of Guanine of A chain and O2 and N3 of Cytosine residue of B chain, the structures of guanine ring is affected and tilt the base pairs of the helix axis. 43, 44 Similarly, the addition of Cu disturbs the hydrogen bonds, tilt of bases occur in adjacent base pairs consequently winding angle of the DNA changes. The intensity of band decreases as the winding angle increases, and increases the twisting of the chain. This inturn affects sugar puckering from anti to syn stacking patterns resulting inchanges in the structural transition from a B-DNA to intermediate comformation of the C-form of DNA and C-form of DNA to altered B-DNA conformation. The obtained data suggest selective interaction of Cu with guanine-cytosine complementary pairs, followed by DNA cross-linking at those sites which are confirmed by docking studies. This study is thus evidenced for strong interaction of Cu and Aβ1-16 with G-C base pairs have a that markedly changes the secondary and tertiary structure of DNA.
The altered B-DNA and C-DNA are energetically weak and are intermediate conformations between B-Z DNA reported in moderately affected AD brain. 44, 45 An energetically weak non-B DNA secondary structures influence the organization of chromosomes and are considered to be involved in both transcriptional activation and inactivation are likely to play a significant role in genomic instability. In addition to the structural changes, the brain is particularly vulnerable to the ROS, resulting in cellular toxicity and decreases the DNA repair mechanism and increase the oxidative DNA damage. Oxidative DNA damage accumulated in the brain increases with age and plays a central role in the pathogenic mechanism of AD. 46, 47 Our work highlights as to how Cu and Aβ are involved in altering DNA topology. The changes in DNA topology will undoubtedly influence the error rates and perhaps the kind of mutations in the promoter regions, which has great relevance to gene expression and activity of transcription factor. Hence, our study supports the idea that differential DNA stability and structural variations in promoter regions cause is a potential pre gains importance in showing the Cu and Aβ1-16 induced DNA damage and neuronal cell death in the early progression of neurodegenerative diseases. The Cu and Aβ interact with of Poly (GC) sequences and inducing conformational changes and stability of DNA.